Our Research
Understanding bacterial evolution, adaptation and pathogenicity through (epi)genomics
About our research
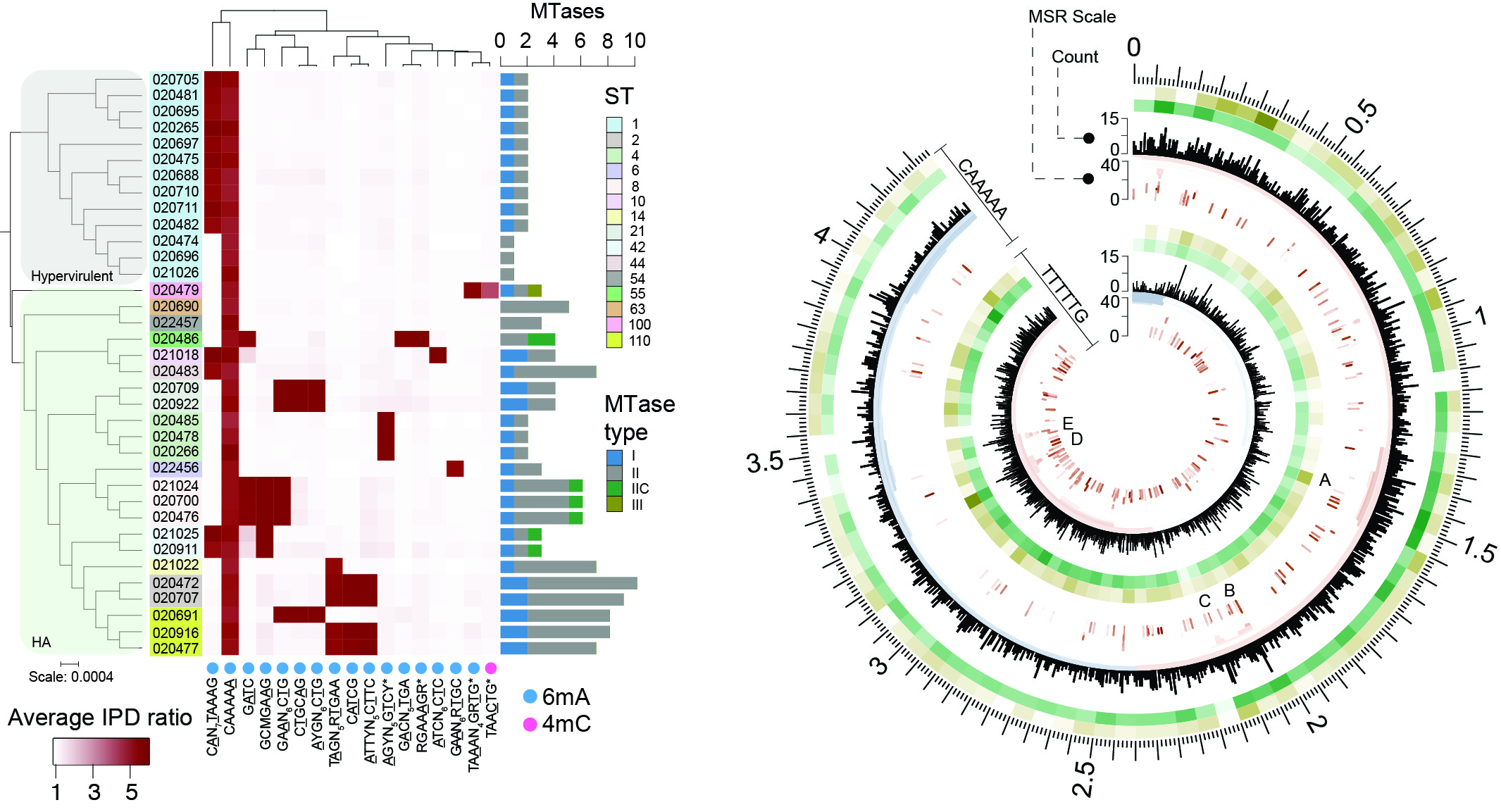
Epigenomics of bacteria
Our ability to comprehensively analyse bacterial epigenomes is facilitated by characterising methylation events and motifs alongside transcriptomic data. We have used SMRT-seq for comparative analysis of pathogens like Clostridioides difficile.
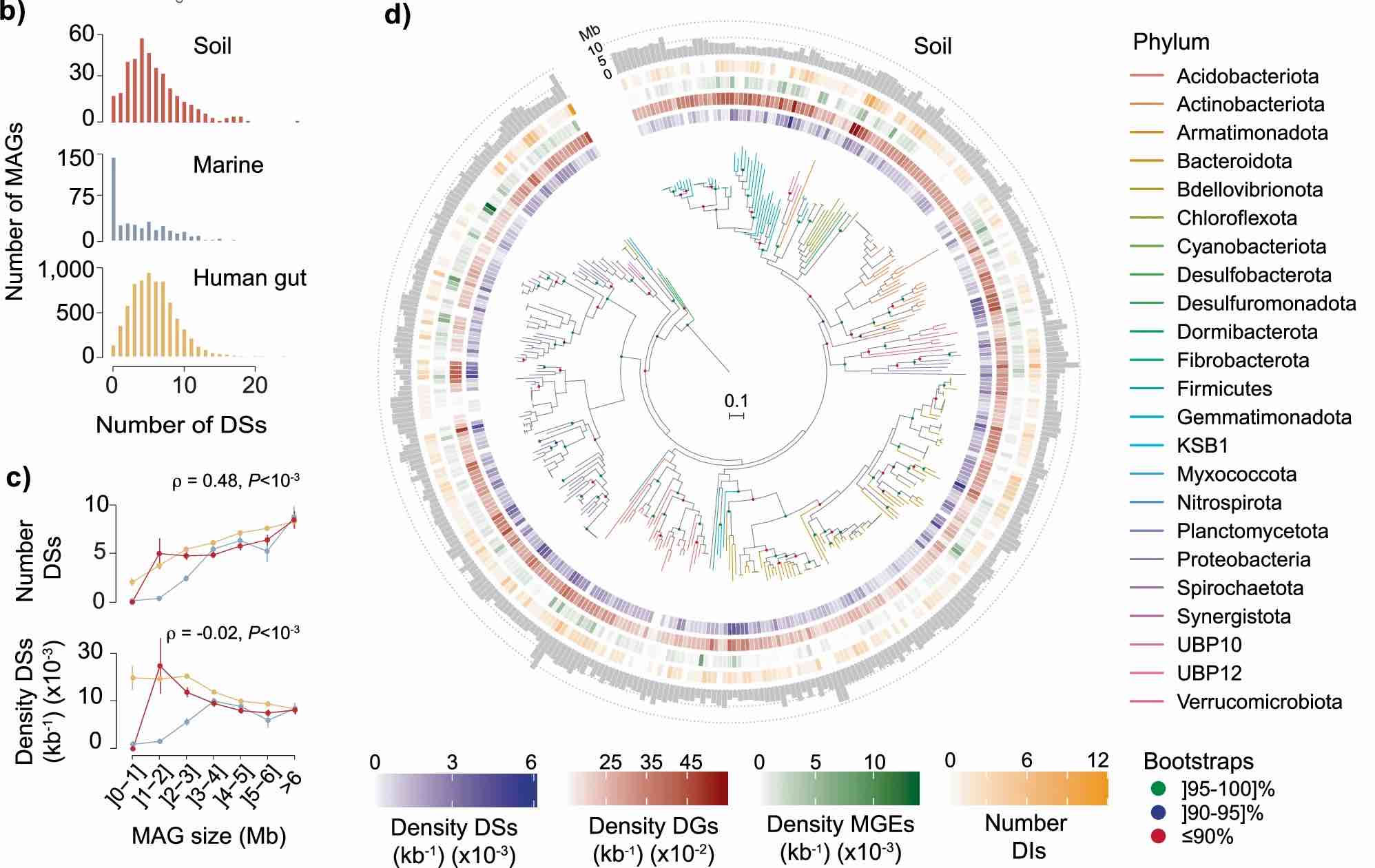
Defensomes and counter-defensomes
Bacteria have developed various defense mechanisms to avoid infection. Our group uses culture-independent metagenomics to advance our understanding of these strategies in microbiota and phageome.
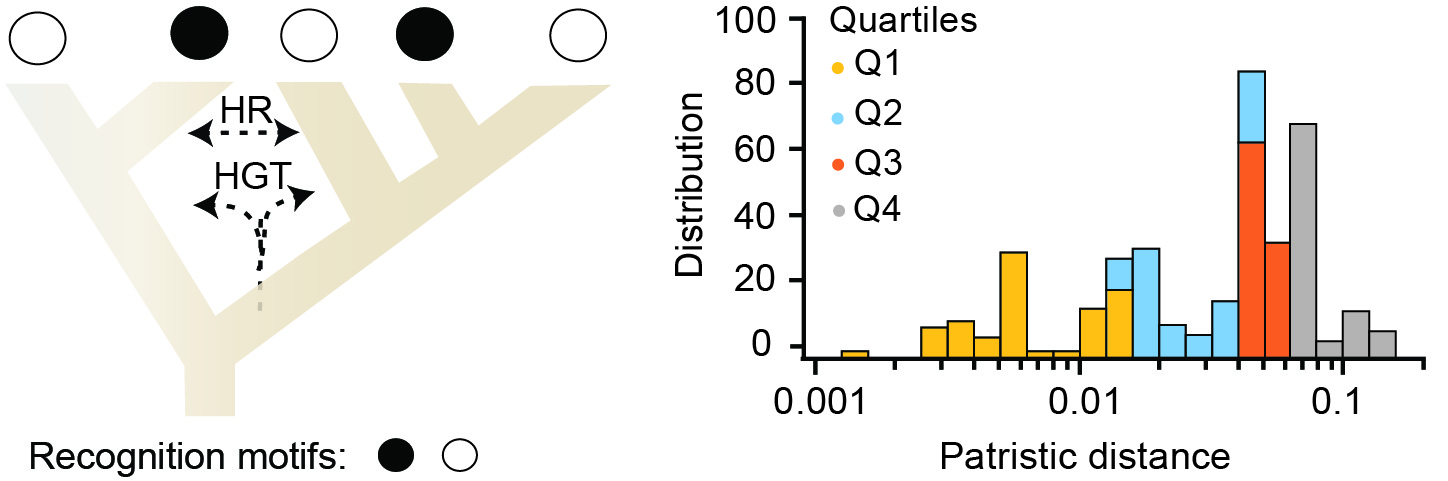
Control of genetic flux by methylation
One of our research interests focuses on the control of genetic flux in bacterial populations by R-M systems. Using comparative genomics we have performed a large-scale analysis of R-M system's abundance, evolution, and association with different mechanisms of genetic mobility.
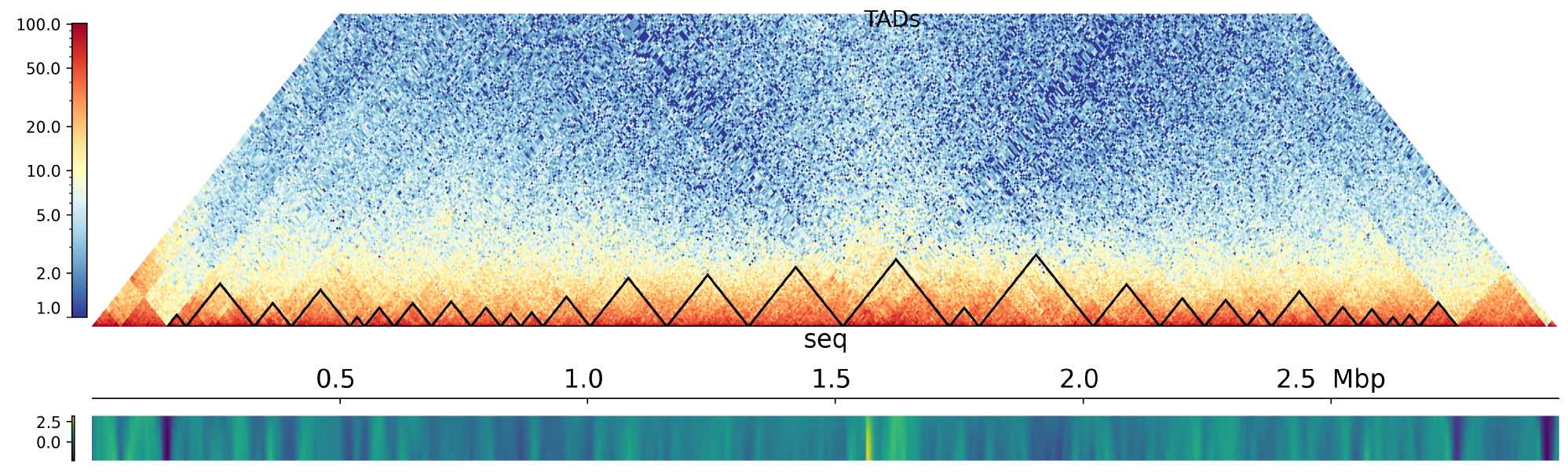
Interplay methylome-transcriptome-DNA topology
Methylation affects chromosome structure and DNA stability. We evaluate how DNA modifications alter the conformation of recognition sites and the resulting expression variability of nearby genes.
To read more about our research, see our publications.
