Laboratoire de Bioinformatique pour la Génomique et la Biodiversité
Bioinformatics Laboratory for Genomics and Biodiversity (LBGB)
La thématique principale de l'équipe est le traitement des données issues des séquençeurs nouvelle génération. Le groupe interagit avec le laboratoire de séquençage, l'équipe développement technologique (développement de nouveaux protocoles et mise en place de nouveaux séquençeurs), et les équipes de recherche. Les missions sont multiples : la mise en forme des données produites par les séquençeurs, le contrôle qualité des données, l'assemblage de génomes et de transcriptomes, l'annotation de génomes eucaryotes.
Our lab is part of the Genoscope and attached to the Institut de Biologie François Jacob of the CEA. The French Alternative Energies and Atomic Energy Commission ( CEA ) is a key player in research, development and innovation in four main areas: defense and security, low carbon energies (nuclear and renewable energies), technological research for industry, fundamental research in the physical sciences and life sciences. Currently the CEA has more than 20,000 employees and is established in nine centers spread throughout France.
Genoscope was founded in 1996 to contribute to the Human genome project and develop genomic programs in France and has subsequently turned toward environmental genomics. It has been part of the CEA Fundamental Research Division (DRF, The Knowledge Factory) since 2006 (Biology field). Genoscope is developing methods and projects for the exploitation of biodiversity, in particular with respect to massive DNA sequencing and bioinformatics. It has been open to the national scientific community through calls for coordinated projects in the context of France Genomique since 2012. The projects cover all biodiversity, particularly the genomics of plants and fungi and the metagenomics of complex ecosystems. Genoscope is affiliated to the Paris-Saclay university which is one of the leading French and European universities, rated 13th in the 2021 Shanghai ranking and recognised for the quality of both its educational programmes and teaching staff. The university also boasts high international visibility thanks to the reputation of its 275 research laboratories and their teams and provides outstanding daily support for the integration and development of 65,000 multicultural students.
Genoscope is composed of several research laboratories with both sequencing and IT equipment. The sequencing laboratory operates short- and long-reads technologies (Illumina, MGI and Oxford Nanopore) and has the capacity to operate large-scale genomic projects with a high number of samples. Since 2012, Genoscope has managed and sequenced the samples from the Tara Ocean expeditions. Genoscope incorporates a 1700-core computing cluster with several large-memory nodes (2-6Tb), and a globally distributed storage of 1.5 PB.In addition, the Genoscope has access to the CEA computing infrastructure. ( CCRT with dedicated large-scale computing infrastructure and storage of 5 PB )
Nos missions
Our mission
The main activities of the LBGB is to develop and evaluate new bioinformatics technologies and software to be used in original and large-scale genomic projects, and in particular with the goals of generating chromosome-scale assemblies of complex genomes based on a combination of long-reads sequencing with long-range information; of providing a gene annotation platform for eukaryotic genomes; and of performing comparative genomics analyzes aiming to establish links between the specificities of a given genome and its life traits. Here are several topics we are currently working on:
Evaluation of sequencing technologies and quality control
In connection with the sequencing lab, we continuously evaluate the sequencing technologies and their associated protocols. This technological survey allows us to propose sequencing strategies adapted to Genoscope projects. We are also developing bioinformatic tools to check the quality of sequencing data produced at Genoscope.
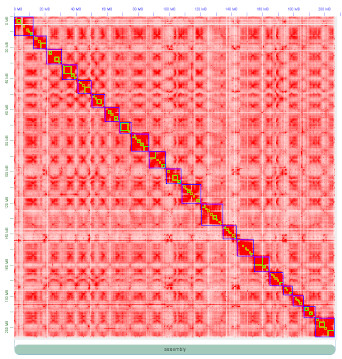
Genome assemblies at the chromosome-scale
Standards are evolving rapidly, and chromosome-level assemblies, as well as annotations integrating state-of-the-art methods are needed. The first sequencing of the Tara Oceans project generated a high proportion of unknown sequences, showing the strong need to generate a more complete database of marine organisms. We are developing new methodologies with the final goal of obtaining near-complete genomes of unknown organisms, presently an unreached goal for eukaryotes.
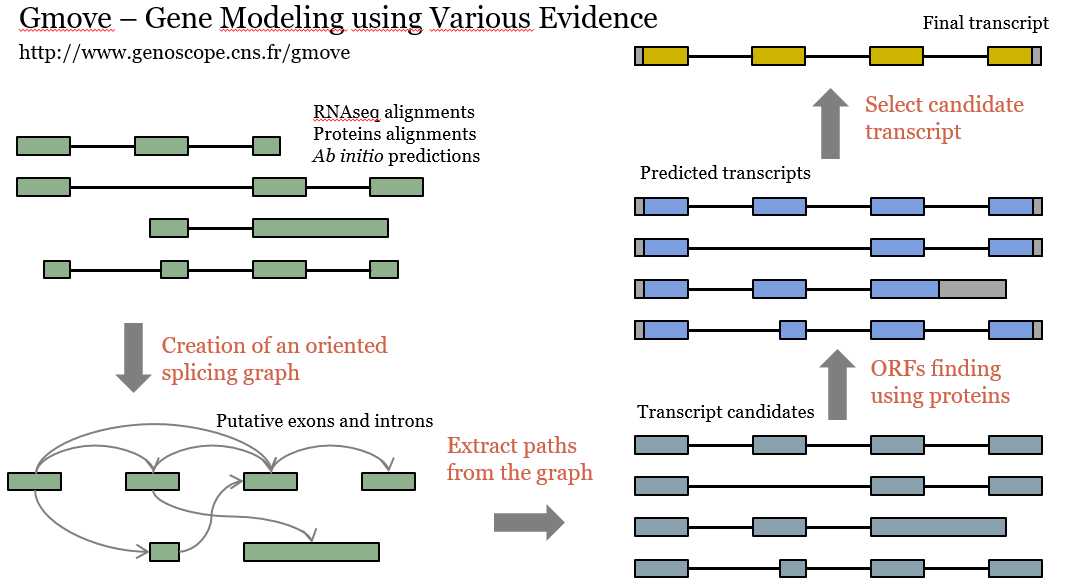
Gene prediction of eukaryotic genomes
Moreover, with the dropoff of the sequencing costs, we could expect that a large variety of genomes will be resequenced, with the goal of generating several references assemblies for a given species. One bottleneck will be the gene prediction, and for this we are working on the development of a gene predictor, called Gmove, that can be used to perform de novo gene prediction as well as to transfer annotations from one genome to another.
Transcriptome profiling
Specific developments in the field of transcriptomics to integrate the new possibilities of nanopore sequencing using RNA molecules. We plan to create tools for building complete transcript maps and their associated expression profiles across experiments, adapted to the environmental datasets produced by the different consortia with whom we are collaborating.
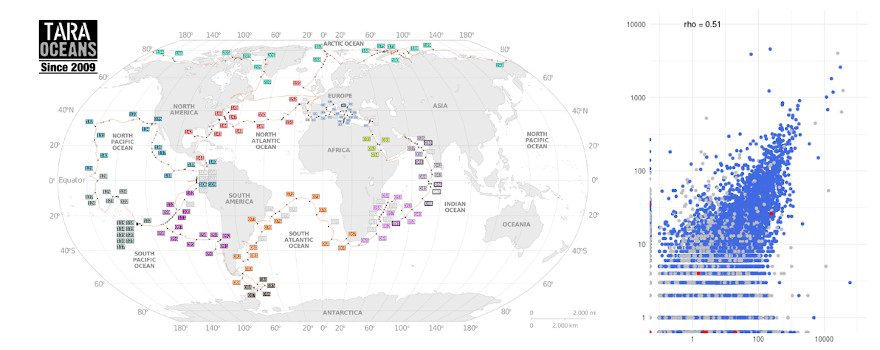
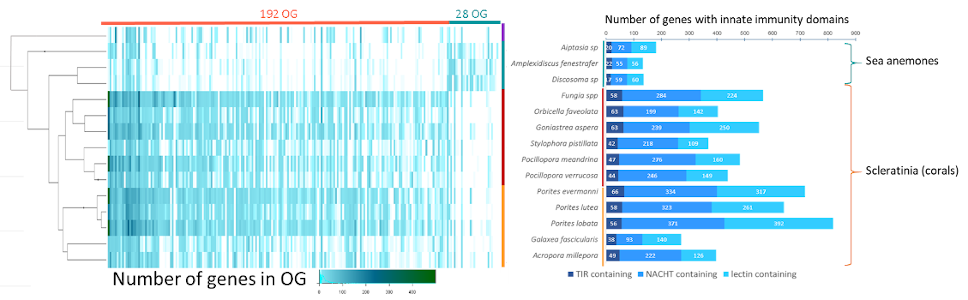
Comparative genomics
Long-lived sessile organisms must persist in the face of a wide range of abiotic and biotic threats over their lifespans. We investigated the genomic features associated with such a long lifespan by sequencing, assembling and annotating genomes of several species. We then used the growing number of whole-genome sequences to investigate the parallel evolution of genomic characteristics potentially underpinning longevity.
Vizualisation
Comparative genomic analysis requires visualization tools, for that purpose, assemblies and genomic features are available through a dedicated interface based on the Generic Genome Browser (GGB, add link). We also develop specific tools that allow us to investigate synteny between genomes and evolutionary history of studied genomes.
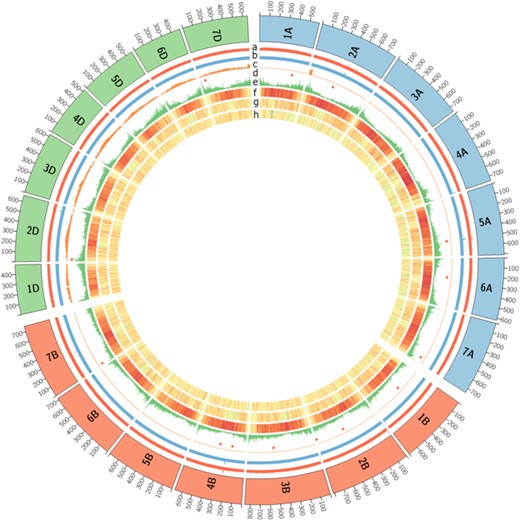
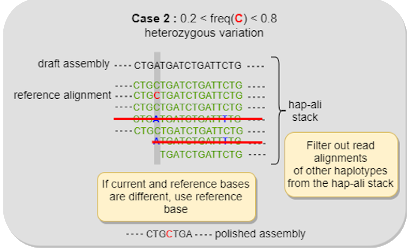
Development of bioinformatics tools
Tools development is guided by the scientific applications of Genoscope. Generally, we use existing software developed by other bioinformatic groups, but we have to manage the bioinformatic issues brought by our own scientific projects. Available tools are not necessarily adapted to our needs, we have to evaluate these tools, modify them and eventually develop new tools.
L'équipe
The team

William AMORY
Production de données de séquençage Sequencing data production

Jean-Marc AURY
Responsable d'équipe Team leader

Caroline BELSER
Assemblage de génome Genome assembly

Arnaud COULOUX
Assemblage de génome Genome assembly

Corinne DA SILVA
Analyse transcriptomique Transcriptomic analysis

France DENOEUD
Génomique comparative Comparative genomics

Lola DEMIRDJIAN
Stagiaire en assemblage de génome Internship on genome assembly

Simone DUPRAT
Assemblage de génome Genome assembly

Phuong DOAN
Prediction de gènes pour le projet BGE Gene prediction for the BGE project

Stéfan ENGELEN
Production de données de séquençage Sequencing data production
Carmen LAFUENTE SANZ
Thèse en metatranscriptomique Thesis on metatranscriptomics

Frédérick GAVORY
Production de données de séquençage Sequencing data production

Benjamin ISTACE
Assemblage de génome Genome assembly

Eléanore LACOSTE
Assemblage de génome Genome assembly

Paul MIELLE
Production de données de séquençage Sequencing data production

Adama NDAR
Assemblage de génome Genome assembly

Benjamin NOEL
Prédiction de g&eagrave;nes Gene prediction

Emilie TEODORI
Stagiaire en assemblage de génome Internship on genome assembly

Marc WESSNER
Prédiction de g&eagrave;nes Gene prediction

Khaoula ZIANE
Prédiction de g&eagrave;nes Gene prediction

Jetez un œil à notre alumni ...
You may consider taking a look at our alumni ...
Most recent and significant publications
Most recent and significant publications
- Integrative omics framework for characterization of coral reef ecosystems from the Tara Pacific expedition. Caroline Belser, Julie Poulain, Karine Labadie, Frederick Gavory, Adriana Alberti, [...], Pedro H. Oliveira, Jean-Marc Aury, Denis Allemand, Serge Planes & Patrick Wincker. Scientific Data volume 10, Article number: 326 (2023).
- Pervasive tandem duplications and convergent evolution shape coral genomes. Benjamin Noël, France Denoeud, Alice Rouan, Carol Buitrago-López, Laura Capasso, [...], Eric Gilson, Didier Zoccola, Patrick Wincker, Christian R. Voolstra & Jean-Marc Aury. Genome Biology volume 24, Article number: 123 (2023).
- Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding. Jean-Marc Aury, Stefan Engelen, Benjamin Istace, Cécile Monat, Pauline Lasserre-Zuber, [...], Etienne Paux, Marion Ranoux, Adriana Alberti, Patrick Wincker, Frédéric Choulet. GigaScience, Volume 11, (2022).
- Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Caroline Belser, Franc-Christophe Baurens, Benjamin Noël, Guillaume Martin, Corinne Cruaud, [...], Jaroslav Doležel, Arnaud Lemainque, Patrick Wincker, Angélique D'Hont & Jean-Marc Aury. Communications Biology volume 4, Article number: 1047 (2021).
- BoardION: real-time monitoring of Oxford Nanopore sequencing instruments. Aimeric Bruno, Jean-Marc Aury & Stefan Engelen. BMC Bioinformatics volume 22, Article number: 245 (2021).
- Hapo-G, haplotype-aware polishing of genome assemblies with accurate reads. Jean-Marc Aury, Benjamin Istace. NAR Genomics and Bioinformatics, Volume 3, Issue 2, (2021).
- Long-read assembly of the Brassica napus reference genome Darmor-bzh. Mathieu Rousseau-Gueutin, Caroline Belser, Corinne Da Silva, Gautier Richard, Benjamin Istace, [...], Jérôme Morice, Patrick Wincker, France Denoeud, Anne-Marie Chèvre, Jean-Marc Aury. GigaScience, Volume 9, Issue 12, (2020).
- BiSCoT: improving large eukaryotic genome assemblies with optical maps. Benjamin Istace, Caroline Belser, Jean-Marc Aury. PeerJ, Volume 8 (2020).
- Transcriptome profiling of mouse samples using nanopore sequencing of cDNA and RNA molecules. Camille Sessegolo, Corinne Cruaud, Corinne Da Silva, Audric Cologne, Marion Dubarry, Thomas Derrien, Vincent Lacroix & Jean-Marc Aury. Scientific Reports volume 9, Article number: 14908 (2019).
- Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Caroline Belser, Benjamin Istace, Erwan Denis, Marion Dubarry, Franc-Christophe Baurens, [...], Mathieu Rousseau-Gueutin, Valérie Barbe, Corinne Cruaud, Patrick Wincker & Jean-Marc Aury Nature Plants volume 4, pages879-887 (2018).
- Oak genome reveals facets of long lifespan. Christophe Plomion, Jean-Marc Aury, Joëlle Amselem, Thibault Leroy, Florent Murat, [...], Francis Martin, Patrick Wincker, Hadi Quesneville, Antoine Kremer & Jérôme Salse. Nature Plants volume 4, pages440-452 (2018).
Nos outils bioinformatiques
Our bioinformatics tools
Stages et Postes
Internships and Positions
Each year, we welcome several students to do their internship in our laboratory. If you are interested in doing your internship in one of our scientific fields, do not hesitate to apply by sending us a CV to: stage_lbgb@genoscope.cns.fr
Job offers:
Offres d'emploi:
Contact
Contact us
Jean-Marc Aury
jmaury@genoscope.cns.fr
01 60 87 25 00









