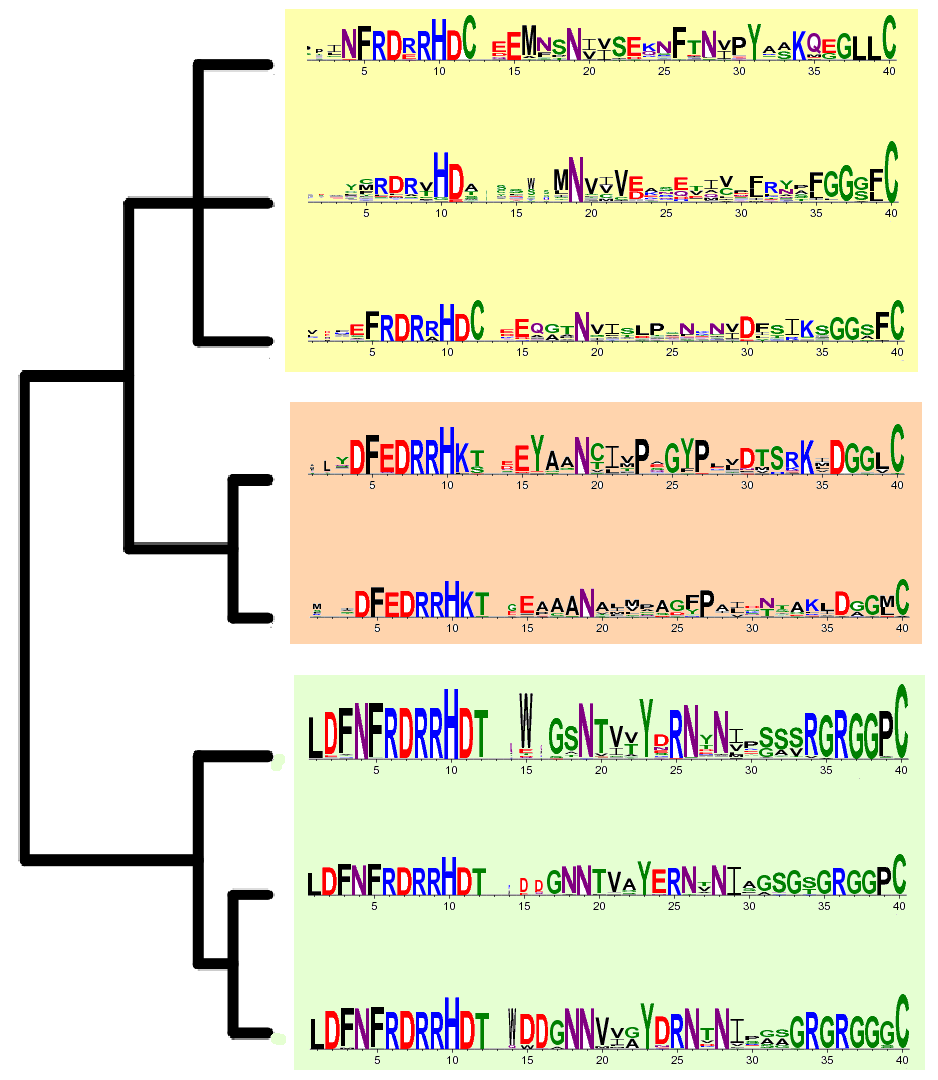
Family PF02274
Amidinotransferase: This family contains glycine (EC:2.1.4.1) and inosamine (EC:2.1.4.2) amidinotransferases, enzymes involved in creatine and streptomycin biosynthesis respectively. This family also includes arginine deiminases, EC:3.5.3.6. These enzymes catalyse the reaction: arginine + H2O <=> citrulline + NH3.