MetX enzymes use succinyl-CoA if an arginine is introduced into one of the two key positions, i.e. P11 or P17, as proved by our active site clustering and our site-directed mutagenesis results. A small fraction of MetX, which does not show up the binding residues of homoserine, cannot carry out the acylation of homoserine. For MetA enzymes, HTS activity is performed by two groups: the first one displays a very similar active site to MetA-HTA enzymes but with a small and neutral residue in P5 while the second one has revealed a specific active site for which the residue in P5 have no influence on the acyl specificity.
MetX-HTA: Download the model of acetylated MetX from Haemophilus influenzae here
MetX-HTS-1: Download the model of succinylated MetX from Acinetobacter baylyi here
MetX-HTS-2: Download the model of succinylated MetX from Frateuria aurantia here
MetA-HTA: Download the model of acetylated MetA from Bacillus cereus here
MetA-HTS-1: Download the model of succylated MetA from Escherichia coli here
MetA-HTS-2: Download the model of acetylated MetA from Methylococcus capsulatus here
MetA and MetX perform identical activities although the catalytic and substrate-binding residues (for L homoserine and acyl-CoA) are different. However, while measuring the distance between the functional groups of the active sites residues, a similar geometry between MetA and MetX catalytic sites came out, as shown in the figure below. Indeed, the nucleophilic base (Cys142 and Ser143 for MetA and MetX, respectively) is actually at a comparable distance from the two amino groups of the peptide bond that stabilize the carbonyl group of the acyl-CoA moiety (i.e. Ala108 plus Phe143 for MetA, and Leu49 plus Phe144 for MetX). Likewise, the functional groups of the residues that interact with L-homoserine (Ser192 plus Arg249 for MetA, and Asp338 plus Arg212 for MetX) form with the catalytic nucleophilic base a triangle with dimensions almost identical in MetA and MetX. In sum, although having active site configuration and composition quite different, MetA and MetX achieve the acetylation/succinylation of homoserine using identical chemical mechanism.
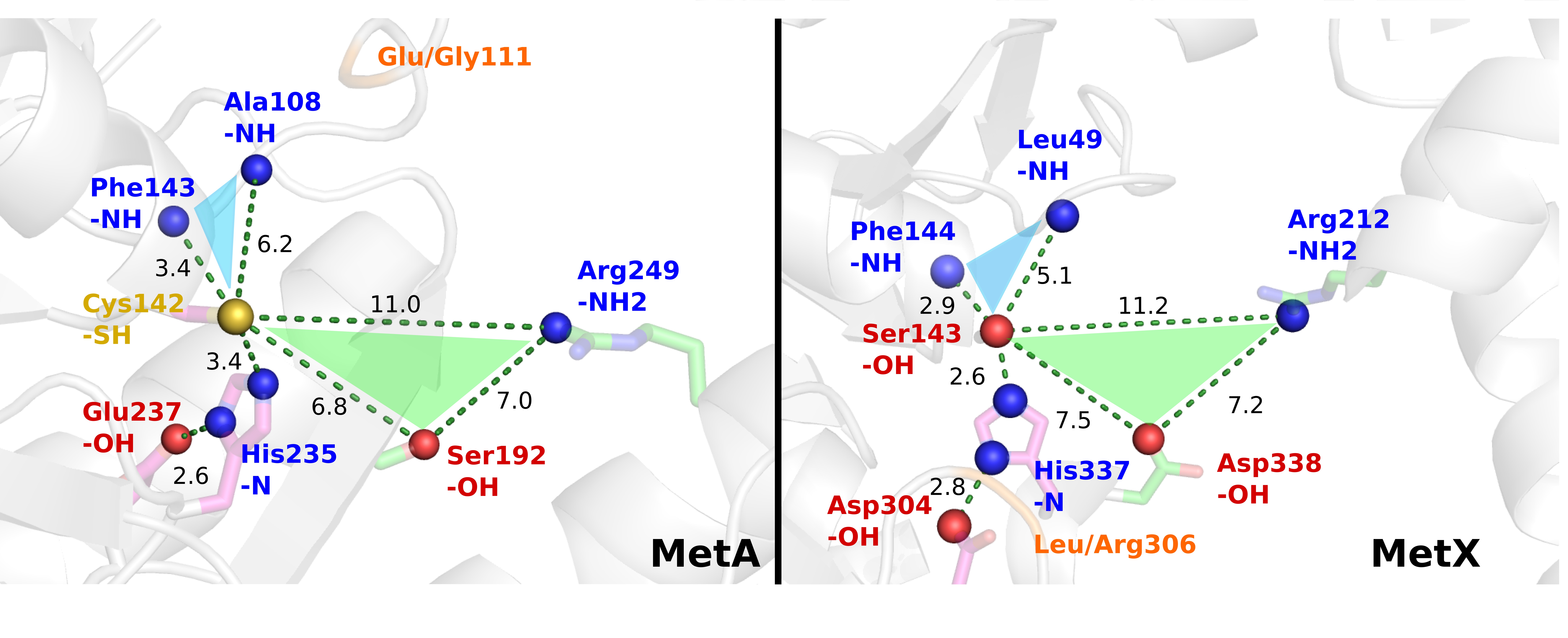
The active site residues involved in catalysis and L-homoserine and acyl-CoA binding are represented in transparent sticks and annotated for MetA cereus (PDB code: 2VDJ) and MetX influenzae (PDB code: 2B61). Their functional groups are represented by a sphere. The blue and green triangles represent the position of the acyl-CoA moiety carbonyl group and the position of the homoserine respectively. The position of the key residues responsible for acyl-CoA specificity is indicated in orange for MetA and for MetX. Distances are indicated in Å.